Data set used by dataquieR and how to import them
How to get and use data sets in dataquieR: reports or single indicator functions
This tutorial informs about how to import study data and metadata to generate data quality reports with dataquieR or to use single indicator functions.
You will need the dataquieR packages installed for this tutorial. Here is the code to install and load it.
# install the package
install.packages("dataquieR")
# load the package library
library(dataquieR)Data used by dataquieR: study data and metadata
Automated data quality assessments using the R package dataquieR work using the collected data (study data), and information and requirements about the study data (metadata).
The functions in dataquieR accept metadata in two formats:
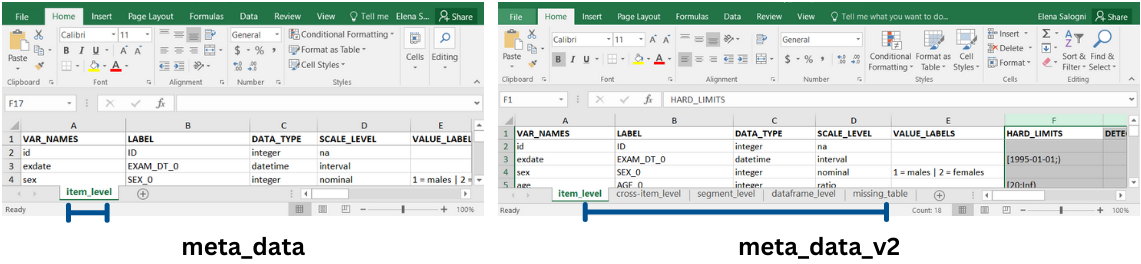
multiple levels metadata: an Excel workbook with multiple spreadsheets containing metadata organized in several tables named following dataquieR conventions: “item_level”, “cross-item_level”, “segment_level”, “dataframe_level”. This type of metadata is identified by the function argument
meta_data_v2 =single level metadata: Each metadata level can be provided individually using a csv file or a single spreadsheet in an Excel file. In this case each argument is specific for the corresponding level as follows: i) item level metadata: a file containing information and expectations about single variables. It is identified by the function argument
meta_data =(alsoitem_level =can be used); ii) cross-item level metadata: file containing information and expectations about groups of variables. It is identified by the function argumentcross-item_level =; iii) segment level metadata: a file containing information and expectations about individual segments (e.g., study examinations). It is identified by the function argumentsegment_level =; iv) dataframe level metadata: a file containing information and expectations about the study data (it can refer to one or more tables). It is indicated by the function argumentdataframe_level =.
See the metadata tutorial for more information.
First case: use dataquieR functions without importing data beforehand
The dq_report2() report generation function and all the
other single indicator functions in dataquieR do not require the study
data and metadata to be uploaded beforehand. Files are directly imported
by the function using a path, a URL,
or just the file name in case of the sample data of the package.
Hereafter there is an example for each case.
1. Use a file path
When you have both the study data (e.g., dataquieR synthetic example data) and the metadata (dataquieR synthetic example metadata) locally available in your “Downloads” folder (click the links to download them), you can create a report using the following example code:
# create a report
# Note that this report will not contain the accuracy dimension
# (To have a complete report add the argument `dimensions = NULL` or `dimensions = "all"`)
rep1 <- dq_report2(study_data = "~/Downloads/study_data_ex1.csv",
meta_data_v2 = "~/Downloads/metadata_ex1.xlsx")
#print the report
rep1
# check range violations of one response variable using the function
# con_limit_deviations()
dev1 <- con_limit_deviations(resp_vars = "SBP_0",
study_data = "~/Downloads/study_data_ex1.csv",
meta_data_v2 = "~/Downloads/metadata_ex1.xlsx")
#print the result in the Viewer
dev12. Use a URL
Data available online can be directly downloaded using the URL to create a report (here is again the synthetic example data directly from the website).
# create a report
rep2<- dq_report2(
study_data = "https://dataquality.qihs.uni-greifswald.de/extdata/study_data.RData",
meta_data_v2 = "https://dataquality.qihs.uni-greifswald.de/extdata/meta_data_v2.xlsx")
rep2
# check the univariate outliers of one response variable using the function
# acc_univariate_outlier()
out1 <- acc_univariate_outlier(
resp_vars = "SBP_0",
study_data = "https://dataquality.qihs.uni-greifswald.de/extdata/study_data.RData",
meta_data_v2 = "https://dataquality.qihs.uni-greifswald.de/extdata/meta_data_v2.xlsx")
#print the result in the Viewer
out13. Use the example data of dataquieR (special case)
Only in the case of the two sample study data and metadata of the dataquieR package (the synthetic example data and the SHIP-based example data), you can specify the name of the files without the path nor the extension.
Here is the example with the SHIP-based example data:
rep3 <- dq_report2(study_data = "ship",
meta_data_v2 = "ship_meta_v2")
rep3In the case of the synthetic example data, you can use the following code:
rep4 <- dq_report2(study_data = "study_data",
meta_data_v2 = "meta_data_v2")
rep44. Use an object in R (data previously imported)
For data already available in the R Global Environment, you can
specify the object names in the dataquieR function you want to use. In
the following example, you will first import the data in R with the name
sd1 for the study data and md1 for the item
level metadata.
#Import example data and metadata in the Global Environment
sd1 <- prep_get_data_frame("study_data")
# You can also import data with other packages, here is
# an example with the "rio" package:
# library(rio)
# sd1 <- import("https://dataquality.qihs.uni-greifswald.de/extdata/study_data.RData")
prep_load_workbook_like_file("meta_data_v2")
md1 <- prep_get_data_frame("item_level")
#Create a report using the objects in R
rep5 <- dq_report2(study_data = sd1,
meta_data = md1)
rep5Second case: use specific dataquieR functions to import data in R
To inspect and modify data is necessary to import them. There are three dataquieR functions for importing data that can be used in different situations:
prep_load_workbook_like_file()prep_load_folder_with_metadata()prep_get_data_frame()
I. Import one metadata Excel Workbook
Multiple levels metadata are Excel workbook files containing multiple spreadsheets named following dataquieR conventions: “item_level”, “cross-item_level”, “segment_level”, “dataframe_level”. For more information see the metadata tutorial.
You can import this type of metadata using the dataquieR function
prep_load_workbook_like_file().
If the metadata is available in a local directory,
e.g., the dataquieR synthetic example metadata file (metadata_ex1.xlsx) that is
present in “Downloads” (click to download it if you did not do it
earlier in the tutorial), you can import the complete Excel file using
the following code:
prep_load_workbook_like_file("~/Downloads/metadata_ex1.xlsx") Otherwise if you are importing the metadata examples of the
dataquieR package (meta_data_v2 or
ship_meta_v2), the dataquieR function
prep_load_workbook_like_file() can be used with the name of
the files without any path or extension:
prep_load_workbook_like_file("meta_data_v2") # for the synthetic example data
prep_load_workbook_like_file("ship_meta_v2") # for the SHIP-based example dataAfter running the prep_load_workbook_like_file()
function, no object will be visible in the Global Environment, as the
data frames will be stored in dataquieR data frame storage area. See the
tutorial Revise
content in the dataquieR data frame storage area for more
information about managing this storage area.
To get the data frames in the Global Environment use the function
prep_get_data_frame.
# To get the item level metadata
md1 <- prep_get_data_frame("item_level")
# To get the cross-item level metadata
cil1 <- prep_get_data_frame("cross-item_level")
# To get the segment level metadata
segl1 <- prep_get_data_frame("segment_level")
# To get the data frame level metadata
df1<- prep_get_data_frame("dataframe_level")II. Import all metadata and study data files from a folder
If you want you can load all the metadata files and the study data
that are inside one folder with one function in dataquieR
prep_load_folder_with_metadata(). This function will upload
all the files in the dataquieR storage area.
For example to load all the metadata files in an imaginary folder called “My_metadata_files” in D:/, you would use the following code:
# To load in the dataquieR storage area all the metadata files in the folder
prep_load_folder_with_metadata("D:/My_metadata_files/")Then you can get the tables in the Global Environment using the
function prep_get_data_frame(), as shown before.
III. Import single data files or parts of them
The function prep_get_data_frame() is a powerful tool
and it can be used to import single files, specific sheets/tables,
single or multiple columns, bookmarks, and using prefix to import data
from other packages.
3.1 Import single data files using a path, a URL, or sample data name
The function can be used to import data providing:
- the path of the file (in case the file is in a local folder),
- the URL of the file,
- just the name of the file for the example data available with the package.
sd0 <- prep_get_data_frame("~/Downloads/study_data_ex1.csv") # see earlier in the tutorial to download this file in your local directory
ship1 <- prep_get_data_frame("https://dataquality.qihs.uni-greifswald.de/extdata/ship.RDS") # the URL to the SHIP-based example data
#Example data available with the package
sd1 <- prep_get_data_frame("study_data") # for the synthetic example data
ship2 <- prep_get_data_frame("ship") # for the SHIP-based example data
md1 <- prep_get_data_frame("meta_data") # for the synthetic example item level metadata
md2 <- prep_get_data_frame("ship_meta") # for the SHIP-based example item level metadata3.2 Import single data files from data frames storage area of dataquieR
Any data frame that is available in the data frames storage area of dataquieR, can be fetched by indicating its name.
# Import the dataquieR metadata example for the synthetic data
# containing multiple metadata sheets
prep_load_workbook_like_file("meta_data_v2") # load the data frames to dataquieR storage area
item_level1 <- prep_get_data_frame("item_level") #get the data frame in an object called item_level1
item_level13.3 Import specific sheets/tables by name or index number
To import a specific sheet from an Excel file you have to append the name after the file name separated by a | symbol, or in alternative you can also use the number of the sheet in the file instead of the name.
The file containing the desired sheet (for example
segment_level) can be:
- in your local directory (for example the dataquieR sythetic example
metadata downloaded earlier in this tutorial and available in your
“Downloads” folder - click here in case you did not download the file
earlier
metadata_ex1.xlsx-)
# Import the segment_level sheet from the synthetic example metadata
# saved in the Downloads folder
# By name
seglev1 <- prep_get_data_frame("~/Downloads/metadata_ex1.xlsx|segment_level")
# By index number
seglev2 <- prep_get_data_frame("~/Downloads/metadata_ex1.xlsx|4")- a previously imported file present in your dataquieR storage area
Attention: This works not only for Excel files, but for all files that feature more than one table (e.g., RData, OpenDocument Spreadsheet - ODS).
To show how it is possible to import tables from an RData files, we first create an RData file.
#Create an RData file with 2 objects:
# a vector
vector1 <- 5:10
# a table
table_ex1 <- data.frame(Numbers = 1:3, Letters = LETTERS[1:3])
save(table_ex1, vector1, file = "table_example.RData")You can get the table from the file “table_example.RData” as follows:
df1 <- prep_get_data_frame("table_example.RData|table_ex1")
df1| Numbers | Letters |
|---|---|
| 1 | A |
| 2 | B |
| 3 | C |
However you can not get the vector from the RData file, because the
function prep_get_data_frames() only works with data
frames.
# v1 is a vector, so the following line should fail
try(v1 <- prep_get_data_frame("table_example.RData|vector1"))## Error in base::tryCatch(base::withCallingHandlers({ :
## File "table_example.RData" did not contain a table (data frame)
## according to 'rio'Note: if only one table is present in the .RData file, there is no need to specify the object name.
#Create an RData file with 2 objects
table_ex2 <- data.frame(Numbers = 1:3, Letters = LETTERS[6:8])
save(table_ex2, file = "table_example_2.RData")
df2 <- prep_get_data_frame("table_example_2.RData")
df2| Numbers | Letters |
|---|---|
| 1 | F |
| 2 | G |
| 3 | H |
3.4 Import specific column/s from a determined sheet in a file or in a table
You can access a specific column or several columns from a file or from a data frame using a combination of | and + symbols.
In the following examples we show how you can get two columns from the segment level of the dataquieR sythetic example metadata downloaded earlier in this tutorial, from the missing code of the same example but directly using its file name, and from the item level using the URL of the example data.
# Import the segment_level sheet from the synthetic example metadata
# saved on the desktop, but only the selected columns STUDY_SEGMENT and SEGMENT_ID_VARS
col1 <- prep_get_data_frame("~/Downloads/metadata_ex1.xlsx|segment_level|STUDY_SEGMENT+SEGMENT_ID_VARS")
col1| STUDY_SEGMENT | SEGMENT_ID_VARS |
|---|---|
| STUDY | v00001 |
| PHYS_EXAM | v00001 |
| LAB | v00001 |
# Import the missing table columns (sheet: missing_table)
col2 <- prep_get_data_frame("meta_data_v2|missing_table|CODE_VALUE+CODE_LABEL")| CODE_VALUE | CODE_LABEL |
|---|---|
| 99980 | Missing - other reason |
| 99981 | Missing - exclusion criteria |
| 99982 | Missing - refusal |
# Import the missing table (sheet: item_level)
col3 <- prep_get_data_frame("https://dataquality.qihs.uni-greifswald.de/extdata/meta_data_v2.xlsx|item_level|VAR_NAMES+LABEL")
col3| VAR_NAMES | LABEL |
|---|---|
| v00000 | CENTER_0 |
| v00001 | PSEUDO_ID |
| v00002 | SEX_0 |
3.5 Import data using predefined sources (e.g., bookmarks)
There are bookmarks for some standardized vocabulary available in dataquieR.
This is the list of all the bookmarks for specific vocabulary available in dataquieR:
## [1] "ICD10GM" "ICD10" "ICPC" "SPAT" "NOMESCO"
## [6] "ATC" "ICD9" "SNOMEDrokan" "SNOMED3" "ICD7"Bookmarks can be indicated using < >
or voc:.
# Import the codes of ICD7
prep_get_data_frame("<ICD7>")
# in alternative you can use `voc:`
prep_get_data_frame("voc:ICD7")| key |
|---|
| 1400 |
| 1401 |
| 1408 |
| 1409 |
| 1410 |
Note: There is the possibility to have custom bookmarks. To do so, execute the following:
prep_add_data_frames(`<>` = data.frame(
voc = c("bookmark1", "bookmark2") ,
url = c("data:datasets|cars", "data:datasets|iris")))
prep_get_data_frame("<bookmark1>")3.6 Import data from other packages
To import data contained in other packages you can: i) import from
their data using the prefix using the prefix data:, ii)
import from their extdata
folder) using the prefix extdata:, or iii) import
specifying the path using the prefix package:.
Let’s say we want to import the data frame iris from the
package datasets. We want to import only 2 columns from
that data frame. This can be done with prep_get_data_frame
using the prefix data: as follows.
# Import the codes of data frame iris from package datasets, only columns
# Sepal.Length and Sepal.Width
prep_get_data_frame("data:datasets|iris|Species+Sepal.Length+Sepal.Width")If we are interested in a data frame that is present in the
extdata of a package, in the folder inst. We
can use the prefix extdata:, as follows.
if (!rlang::is_installed("tor")) {
install.packages("tor")
}
prep_get_data_frame("extdata:tor/csv/csv1.csv")Finally , to access to the data frame we need to indicate all the
path to the specific data frame and the prefix package:.
For example for the package dataquieR we should write as follows.
prep_get_data_frame("package:tor/extdata/csv/csv1.csv")


