Data type mismatch
The Data
type mismatch indicator be calculated using int_datatype_matrix
in the following way:
# Load dataquieR
library(dataquieR)
# Load data
sd1 <- prep_get_data_frame("ship")
# Load metadata
prep_load_workbook_like_file("ship_meta_v2")
meta_data_item <- prep_get_data_frame("item_level") # item_level is a sheet in ship_meta_v2.xlsx
# Apply indicator function
datatype_res <- int_datatype_matrix(
study_data = sd1,
meta_data = meta_data_item,
label_col = "LONG_LABEL"
)A plot and a table are provided to view the results:
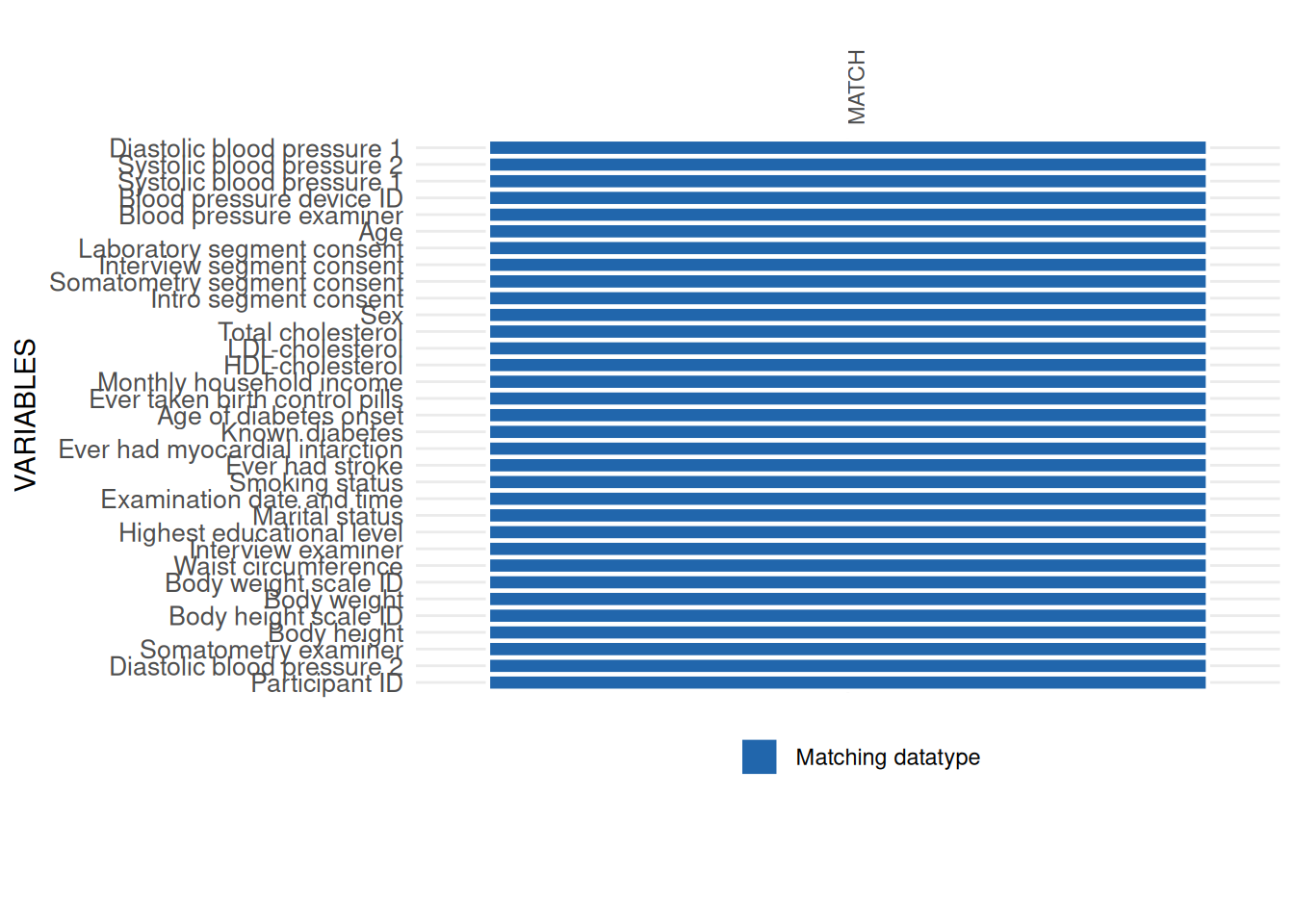
datatype_res$SummaryPlot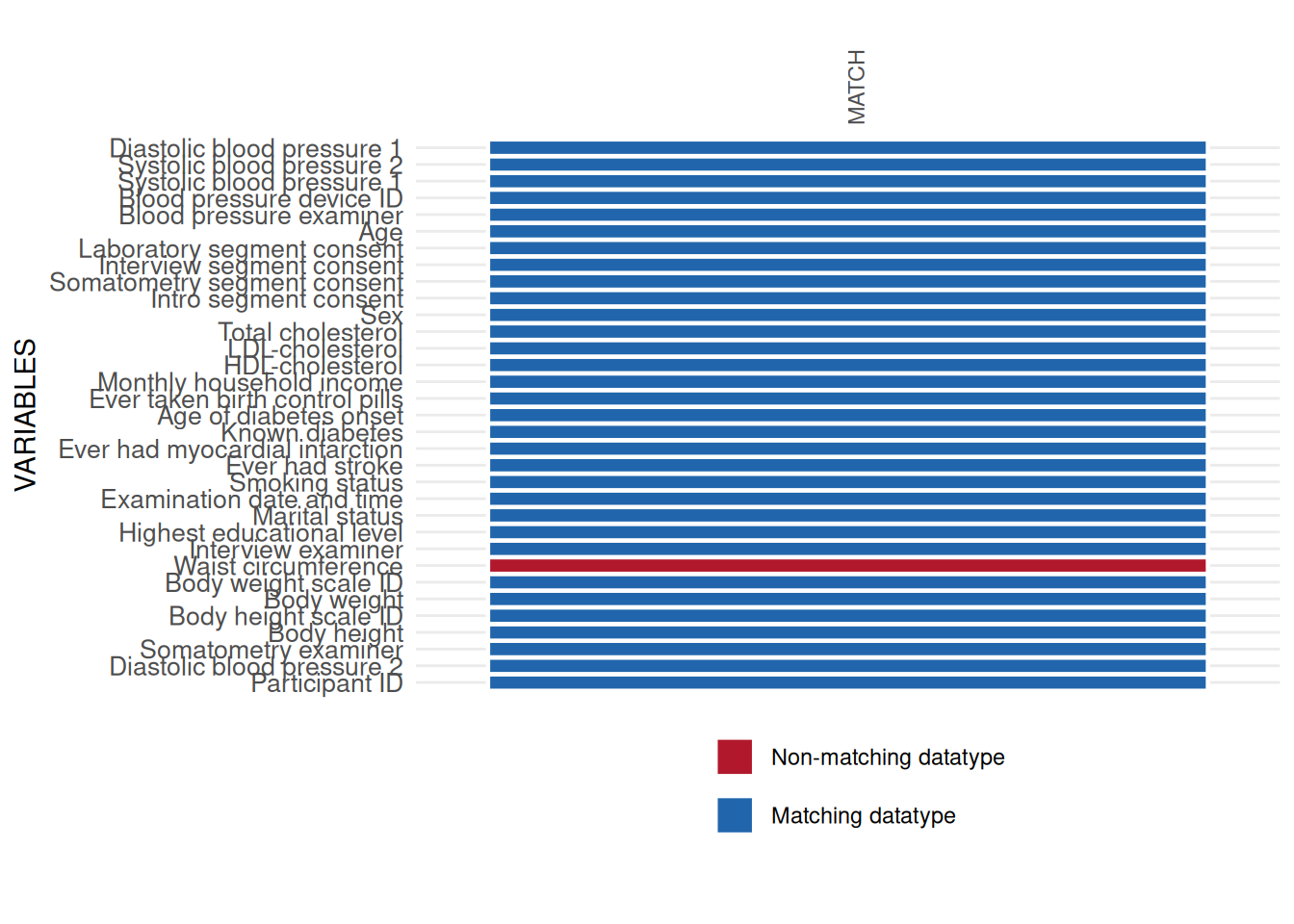
datatype_res$SummaryData| Variables | Data type mismatch N (%) | Convertible mismatch, stable N (%) | Convertible mismatch, unstable N (%) | Data type match N (%) | Expected DATA_TYPE | Observed DATA_TYPE | State, given threshold | STUDY_SEGMENT | |
|---|---|---|---|---|---|---|---|---|---|
| 25 | Participant ID | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 10 | Diastolic blood pressure 2 | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 28 | Somatometry examiner | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 5 | Body height | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 6 | Body height scale ID | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 7 | Body weight | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | float | float | Matching datatype | SOMATOMETRY |
| 8 | Body weight scale ID | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 33 | Waist circumference | 3 (0.14%) | 2148 ( 99.72%) | 0 (0.00%) | 3 ( 0.14%) | float | string | Non-matching datatype | SOMATOMETRY |
| 17 | Interview examiner | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 16 | Highest educational level | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 23 | Marital status | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 14 | Examination date and time | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | datetime | datetime | Matching datatype | INTRO |
| 27 | Smoking status | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 12 | Ever had stroke | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 11 | Ever had myocardial infarction | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 20 | Known diabetes | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 2 | Age of diabetes onset | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 13 | Ever taken birth control pills | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 24 | Monthly household income | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTERVIEW |
| 15 | HDL-cholesterol | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | float | float | Matching datatype | LABORATORY |
| 22 | LDL-cholesterol | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | float | float | Matching datatype | LABORATORY |
| 32 | Total cholesterol | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | float | float | Matching datatype | LABORATORY |
| 26 | Sex | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 19 | Intro segment consent | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 29 | Somatometry segment consent | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 18 | Interview segment consent | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 21 | Laboratory segment consent | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 1 | Age | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | INTRO |
| 4 | Blood pressure examiner | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 3 | Blood pressure device ID | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 30 | Systolic blood pressure 1 | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 31 | Systolic blood pressure 2 | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
| 9 | Diastolic blood pressure 1 | 0 (0.00%) | 0 ( 0.00%) | 0 (0.00%) | 2154 (100.00%) | integer | integer | Matching datatype | SOMATOMETRY |
All datatype issues found by int_datatype_matrix
should be checked data element by data element. For instance, a major
issue was found in the variable WAIST_CIRC_0. This variable is
in the study data with datatype character, which differs from
the expected datatype float defined in the metadata. Some basic
checks show the misuse of commas as the decimal delimiter.
int_inspect_char(sd1$waist)| Character | Count |
|---|---|
| , | 3 |
| . | 2144 |
| 0 | 933 |
| 1 | 1443 |
| 2 | 908 |
| 3 | 884 |
| 4 | 889 |
| 5 | 898 |
| 6 | 1018 |
| 7 | 1279 |
| 8 | 1355 |
| 9 | 1409 |
| NA | 3 |
To correct this issue, converting WAIST_CIRC_0 to datatype
numeric will coerce respective values to NA’s,
which should be avoided. Hence, we replace the comma with the correct
delimiter and correct the datatype without losing data values. The
resulting applicability plot shows no more issues.
# replace comma with the correct delimiter
sd1$waist <- as.numeric(gsub(",", ".", sd1$waist))
int_datatype_matrix(
study_data = sd1,
meta_data = meta_data_item,
label_col = "LONG_LABEL"
)$SummaryPlot