Item missingness
Uncertain missingness status, Missing due to specified reason, and Missing values can be assessed using com_item_missingness
as in the following call:
# Load dataquieR
library(dataquieR)
# Load data
sd1 <- prep_get_data_frame("ship")
# Load metadata
prep_load_workbook_like_file("ship_meta_v2")
meta_data_item <- prep_get_data_frame("item_level") # item_level is a sheet in ship_meta_v2.xlsx
# Apply indicator function
item_miss <- com_item_missingness(
study_data = sd1,
meta_data = meta_data_item,
show_causes = TRUE,
label_col = "LABEL",
include_sysmiss = TRUE,
threshold_value = 95
) An overview can be obtained by calling a summary table:
item_miss$SummaryTable| Variables | Expected observations N | Sysmiss N | Datavalues N | Missing codes N | Jumps N | Measurements N | Missing_expected_obs | PCT_com_crm_mv | GRADING |
|---|---|---|---|---|---|---|---|---|---|
| ID | 2154 | 0 | 2154 | 0 | 0 | 2154 | 0 | 0.00 | 0 |
| EXAM_DT_0 | 2154 | 0 | 2154 | 0 | 0 | 2154 | 0 | 0.00 | 0 |
| SEX_0 | 2154 | 0 | 2154 | 0 | 0 | 2154 | 0 | 0.00 | 0 |
| AGE_0 | 2154 | 0 | 2154 | 0 | 0 | 2154 | 0 | 0.00 | 0 |
| OBS_BP_0 | 2154 | 0 | 2154 | 3 | 0 | 2151 | 3 | 0.14 | 0 |
| DEV_BP_0 | 2154 | 0 | 2154 | 3 | 0 | 2151 | 3 | 0.14 | 0 |
| SBP_0.1 | 2154 | 2 | 2152 | 0 | 0 | 2152 | 2 | 0.09 | 0 |
| SBP_0.2 | 2154 | 6 | 2148 | 0 | 0 | 2148 | 6 | 0.28 | 0 |
| DBP_0.1 | 2154 | 2 | 2152 | 0 | 0 | 2152 | 2 | 0.09 | 0 |
| DBP_0.2 | 2154 | 6 | 2148 | 0 | 0 | 2148 | 6 | 0.28 | 0 |
| OBS_SOMA_0 | 2154 | 0 | 2154 | 2 | 0 | 2152 | 2 | 0.09 | 0 |
| BODY_HEIGHT_0 | 2154 | 3 | 2151 | 0 | 0 | 2151 | 3 | 0.14 | 0 |
| DEV_HEIGHT_0 | 2154 | 0 | 2154 | 2 | 0 | 2152 | 2 | 0.09 | 0 |
| BODY_WEIGHT_0 | 2154 | 4 | 2150 | 0 | 0 | 2150 | 4 | 0.19 | 0 |
| DEV_WEIGHT_0 | 2154 | 0 | 2154 | 2 | 0 | 2152 | 2 | 0.09 | 0 |
| WAIST_CIRC_0 | 2154 | 3 | 2151 | 0 | 0 | 2151 | 3 | 0.14 | 0 |
| OBS_INT_0 | 2154 | 0 | 2154 | 1 | 0 | 2153 | 1 | 0.05 | 0 |
| SCHOOL_GRAD_0 | 2154 | 0 | 2154 | 113 | 0 | 2041 | 113 | 5.25 | 1 |
| RELATION_STATUS_0 | 2154 | 0 | 2154 | 67 | 0 | 2087 | 67 | 3.11 | 0 |
| SMOKING_STATUS_0 | 2154 | 0 | 2154 | 68 | 0 | 2086 | 68 | 3.16 | 0 |
| STROKE_YN_0 | 2154 | 0 | 2154 | 70 | 0 | 2084 | 70 | 3.25 | 0 |
| MYOCARD_YN_0 | 2154 | 0 | 2154 | 74 | 0 | 2080 | 74 | 3.44 | 0 |
| DIABETES_KNOWN_0 | 2154 | 0 | 2154 | 7 | 0 | 2147 | 7 | 0.32 | 0 |
| DIAB_AGE_ONSET_0 | 2154 | 0 | 2154 | 7 | 1974 | 173 | 1981 | 91.97 | 0 |
| CONTRACEPTIVA_EVER_0 | 2154 | 0 | 2154 | 4 | 1264 | 886 | 1268 | 58.87 | 0 |
| HOUSE_INCOME_MONTH_0 | 2154 | 0 | 2154 | 116 | 0 | 2038 | 116 | 5.39 | 1 |
| CHOLES_HDL_0 | 2154 | 16 | 2138 | 0 | 0 | 2138 | 16 | 0.74 | 0 |
| CHOLES_LDL_0 | 2154 | 28 | 2126 | 0 | 0 | 2126 | 28 | 1.30 | 0 |
| CHOLES_ALL_0 | 2154 | 15 | 2139 | 0 | 0 | 2139 | 15 | 0.70 | 0 |
| SEG_PART_INTRO | 2154 | 0 | 2154 | 2154 | 0 | 0 | 2154 | 100.00 | 1 |
| SEG_PART_SOMATOMETRY | 2154 | 0 | 2154 | 2154 | 0 | 0 | 2154 | 100.00 | 1 |
| SEG_PART_INTERVIEW | 2154 | 0 | 2154 | 2154 | 0 | 0 | 2154 | 100.00 | 1 |
| SEG_PART_LABORATORY | 2154 | 0 | 2154 | 2154 | 0 | 0 | 2154 | 100.00 | 1 |
The table provides one line for each of the 29 variables. Of particular interest are:
- System missings (Sysmiss): the number and percentage of data fields for each variable without any valid data entry, indicating a non-informative missing value (Uncertain missingness status).
- Missing Codes: the number and percentage of data fields with valid missing codes.
- Jumps: the number and percentage of data fields for which no data collection was attempted.
- Measurements: provides an inverse of Missing values.
The table shows that the variable HOUSE_INCOME_MONTH_0
(monthly net household income) is affected by many missing values. In
addition, the age of diabetes onset (DIAB_AGE_ONSET_0) was
only coded for 173 subjects, but most values are missing because of an
intended jump.
Note: In case jump codes have been used, e.g., for
the variable CONTRACEPTIVE_EVER_0, the denominator for
calculating item missingness is corrected for the number of jump codes
used.
The summary plot delivers a different view of missing data by providing the frequency of the specified reasons for missing data.
r <- print(item_miss$ReportSummaryTable, view = FALSE)
r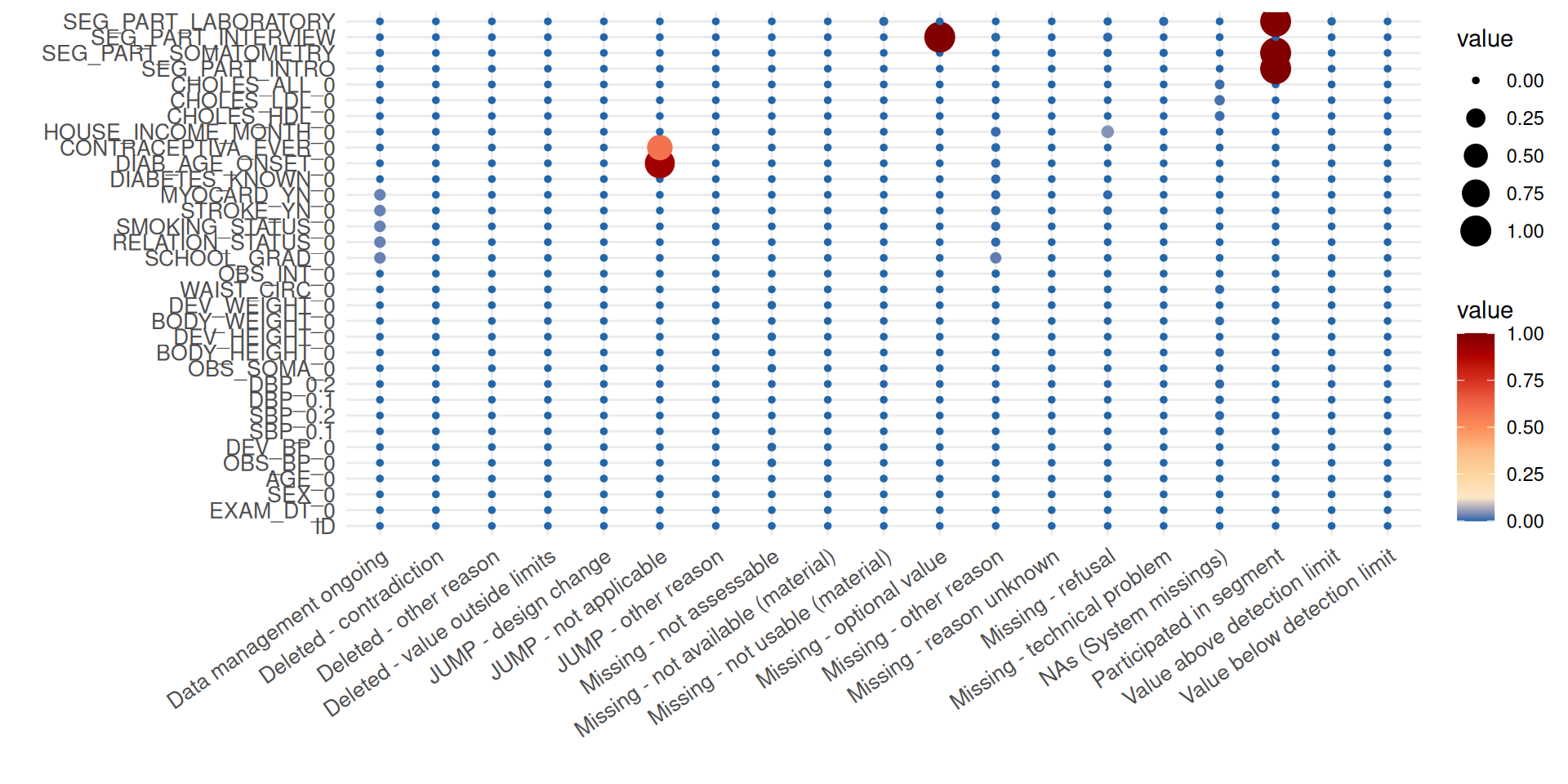
In the plot, the balloon size is determined by the number of missing
data fields. It can now be inferred that, for example, the elevated
number of missing values for the item HOUSE_INCOME_MONTH_0
is mainly caused by refusals of participants to answer the respective
question.



